Search box
Microbiology Concept Inventory
Please note, you must be an educator in higher ed or maybe high school to qualify to recieve the MCI
( 96176 Reads)
|The Enterobacteriaceae are a large group of gram-negative, non-sporulating, short rods that are motile by peritrichous flagella or are non-motile. They have relatively simple nutritional requirements and are facultative anaerobes that ferment glucose to acid under anaerobic conditions. Many of them can be isolated from the intestinal tract and the term enterics has come to be synonymous with the Enterobacteriaceae. However, most are not limited to this environment. The medical importance of many of the members of this family has resulted in intense study of the group. This is probably the best characterized collection of prokaryotes.
Enterics are actually very similar to one another, sharing many biochemical and physiological properties. (Some members can even be attacked by the same bacteriophage.) An important differentiating characteristic of the Enterobacteriaceae is the method used to ferment glucose. Mixed acid fermenters metabolize glucose anaerobically to lactic acid, succinic acid, acetic acid, formic acid, ethanol, CO2, and H2 (see page 17). The large amount of acid produced in this fermentation, lowers the pH of the medium (below pH 4.4). Genera of Enterobacteriaceae that have this type of metabolism include: Proteus, Citrobacter, Edwardsiella, Salmonella, Shigella, and Escherichia.
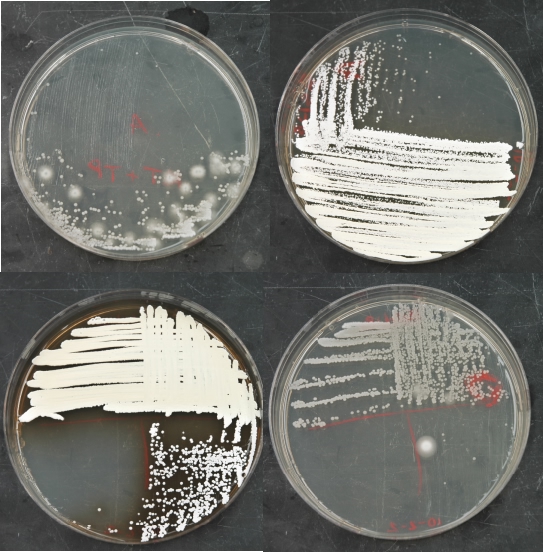
Figure 10.8. Streaks for isolated colonies on penassay agar. The appearance of penassay agar after streaking for isolated colonies of Actinomycetes.
The second groups, those that produce butanediol during fermentation, breaks down glucose into the products mentioned above plus butanediol. Less net acid is produced, due to the formation of a majority of neutral end-products. Eventually, a higher pH (> 6.2) is produced. Klebsiella, Serratia, and Enterobacter have this type of metabolism. Two simple tests, the methyl red test and the Vogues-Proskauer test, have been developed to distinguish these two types of fermentations. Due to the similarity between many genera of Enterobacteriaceae, these two tests are obviously not sufficient to identify them and many more analyses need to be performed. This experiment will introduce several of these tests and how to interpret their results.
A short survey of the major genera
Below are listed a few interesting and relevant facts about some of the Enterobacteriaceae to help familiarize you with this important group. Much more is known about these organisms than what can be presented in this book!
The ability of Salmonella and Shigella to cause disease in humans first sparked interest in the enterics. However, information obtained from studying these organisms has had great impact on our understanding of biological processes in general. An initial step in the disease process is colonization of the intestinal tract. Subsequent pathologies range from discomforting, but relatively mild gastroenteritis, to potentially fatal typhoid fever. Shigella is a close relative of E. coli and most isolates are pathogenic. This organism causes a diarrheal illness that involves colonization and attack of the intestines. One to four days after ingestion the victim will experience a sudden onset of abdominal cramps, fever and profuse bloody diarrhea containing mucous. The illness normally lasts a few days and can be caused by as few as 100 organisms. Improperly prepared food and contaminated water are typical sources of infection with Shigella.
Salmonella is a genus of pathogenic organisms infecting humans and many mammals, birds and reptiles. Organisms in this genus are so genetically similar that they are now considered as belonging to two species S. bongori and S. enterica. S. enterica has six subspecies and each subspecies has its own set of serotypes. Each serotype is distinguished by a unique combination of cell wall and flagellar antigens. Serotype recognition is important in epidemiology; an outbreak or epidemic caused by organisms of one serotype can often be traced to a common source. Decades ago, it was the fashion to consider each serotype a species. Today, most serotypes are given names which are designated as a species names such as Salmonella typhimurium , S. enteritidis and S. heidelberg , but this is only for convenience and is not taxonomically accurate. These three serotypes are responsible for about half of the cases of human Salmonella infection in the United States.
Most serotypes of Salmonella cause gastroenteritis of varying degrees or severity, with or without bacteremia. The source of gastroenteritis is usually contaminated food containing >106 cells/g (or ml). Symptoms generally appear in 8 to 30 hours after ingestion and include nausea, fever, diarrhea, and abdominal pain. Symptoms usually subside in one to two days. Certain host-adapted, biochemically-distinct serotypes cause life-threatening illnesses such as typhoid fever, "hog cholera" and "fowl typhoid".
Organisms of the genus Edwardsiella are known to cause disease in humans as well and a variety of warm and cold-blooded vertebrates. E. tarda is an occasional opportunistic pathogen for humans, causing wound infections and, in less-industrialized countries, gastroenteritis. E. tarda and E. ictaluri have caused massive infections of commercially-raised catfish with considerable economic loss.
The genus Yersinia is an infamous member of the enterics. Y. enterololitica is involved in many cases of gastroenteritis. Y. pestis is the cause of bubonic plague, a dreaded disease until relatively recently. In the fourteenth century the black death, as it was called, claimed the lives of one-third of Europe's population. The disease, in some forms, can kill in a matter of hours. Bubonic plague is harbored in rats and is spread to humans by fleas. Due to good sanitation (a low population of rats) a recurrence of this disease on a large scale in the U. S. is unlikely
Proteus, Providencia and Morganella. This group of non-pathogenic organisms is distinguished by the ability to deaminate phenylalanine. They are often implicated in spoilage of fish and seafood. Proteus forms unusual swarming patterns on agar plates that have a concentric ring shape. This unusual pattern is caused by alternating phases of rapid motility and less motile growth.
Figure 10.9. An appropriately grown test plate. Example plates after growth of the Streptomyces test strains. Note the full growth completely along the streak line. There is a minor contaminant in the left sample, but it should not interfere with analysis.
Citrobacter is a commonly isolated, non-pathogenic enteric. It is of interest because it frequently gives false positives in tests designed to detect the presence of Salmonella in food. Erwinia species are pathogenic for plants causing various soft rots and wilts of important crops and vegetables. If you have ever thrown away soft rotten potatoes, they were probably attacked by E. carotovora, the cause of potato soft-rot.
Occasional strains of Serratia form red pigmented colonies on agar plates. The red color is due to the synthesis of a series of linear tripyroles (prodigiosins). The pigment is curious in that it bears resemblance to the tetrapyroles involved in energy transfers (i.e. hemes, cytochromes, chlorophylls, etc.). The function of the prodigiosins, however is unknown.

Figure 10.10. Antimicrobial production by isolates. Examples of microbes that have been isolated and their inhibition of test microbes. Isolate 3 is secreting a brown metabolite into the medium. Isolate 4 is secreting an antimicrobial into the medium that is inhibiting S. epidermidis, E. faecalis, and B. subtilis. Ec - E. coli, Pf - P. flourescens, Bs - B. subtilis, Se - S. epidermidis, and Ef - E. faecalis.
Coliforms
A coliform is defined as a non-spore-forming, facultatively anaerobic, gram-negative rod, which ferments lactose to acid and gas within 48 hours at 35°C. This is an operational definition used in water analysis (see below) and any organism isolated meeting these requirements is a coliform. In practice, isolated coliforms are almost always Enterobacteriaceae from the genera Enterobacter, Klebsiella, and Escherichia (also lactose positive strains of Citrobacter).
Klebsiella are of interest due to the ability of most strains to use N2 as sole nitrogen source. Study of this organism has helped to unlock many of the mysteries of nitrogen fixation (an agronomically important process). K. pneumoniae is a human pathogen sometimes causing pneumonia. Enterobacter and Klebsiella are widely distributed in water, soil, and plant material.
One member of the genus Escherichia, E. coli, is the star organism of biology. It is also useful as a tool for harboring and amplifying DNA in genetic engineering. This organism's main habitat is the intestinal tract of warm blooded animals, but it can also be found in environments contaminated with feces. Some strains of E. coli also cause gastrointestinal disease. Several recent severe outbreaks have been traced to undercooked meat infected with pathogenic E. coli.
In this experiment we examine the isolation of an enteric from the environment. Since these organisms are everywhere, any natural sample you choose will likely contain enteric organisms. The second half of this experiment involves the analysis of water for fecal contamination. Enteric organisms play an important role in this process.
Water Analysis
Water, the universal solvent, is essential to life. In drier climes, people will even fight over it (look in a newspaper and read about water rights in Colorado and California). Critical to our modern civilization is the availability of a clean water supply for bathing, drinking and cooking. Unfortunately, many pathogens are transmitted through the water supply. Some of these disease-causing pests enter water from the feces of ill individuals and are then ingested and thereby transmitted to others. Diseases such as polio, typhoid, cholera, hepatitis, shigellosis, salmonellosis and others can spread in this manner. To assure a safe water supply, it is critical to monitor for the presence of these pathogens. However, it would be expensive and time consuming to check the water supply for all of them, instead, an indicator organism is used to assay for fecal contamination. Indicator organisms must have four properties to be useful for water analysis.
- The only natural environment of the microbe should be in association with feces and it should always be present.
- It should not grow outside of its natural environment.
- The bacterium should survive longer than the most viable pathogen, but not so long so that historical events are detected.
- It should be easy to detect.
Coliforms come closest to fulfilling all these criteria and are the standard indicator organisms used to test for the biological pollution of water. Enterobacter and Klebsiella are able to survive and multiply in the environment and are therefore not the best indicators of fecal pollution. The sole habitat of E. coli and K. pneumoniae, termed fecal coliforms, is the intestines of warm blooded animals. Thus, fecal coliforms are good indicators of fecal pollution and can be differentiated from other coliforms by incubating on selective media at 44.5°C.
Using coliforms, the EPA has developed standards for clean, safe water. These standards vary, depending upon the waters intended use. Drinking water and the water in swimming pools must be of the highest purity. There can be no more than one positive sample (>1 coliform/100 ml) in 40 samples tested in a month and the concentration of fecal coliforms must be zero. But wait, we just said that some coliforms are present in the environment. How can these standards be met? In good quality well water most microorganisms are filtered out as the water percolates from the surface to the well. Unusually high numbers of coliforms in well water may indicate run-off from a polluted area. In the case of surface waters (rivers and lakes), filtration through a sand bed and chlorination remove most microbes. There may also be further steps that need to be taken to insure water safety depending on the treatment plant. Swimming pools, being open surface water, are often contaminated by organisms in the air or by swimming, bacteria-infested humans. Chlorine is added to keep numbers low. Note that the number of permissible coliforms is not zero. This would be difficult to achieve and would provide no additional gain in safety.
Natural bathing beaches and treated sewage are assayed for numbers of fecal coliforms. Total coliform counts are not used as a measure due to the near ubiquitous presence of Enterobacter and Klebsiella in the environment. If the count reaches > 400 fecal coliforms per 100 ml or a monthly geometric average of > 200 per 100 ml, it may indicate a problem in the sewage treatment process or that the beach should be closed. The latter often happens in heavily utilized beaches during the summer.
Testing for coliforms
Presently, several tests are in use to assay for coliforms in water, The oldest of these is the multiple tube fermentation test. In this test three steps are performed; the presumptive, confirmed, and completed tests. A moderately selective lactose broth medium (Lactose Lauryl Tryptose Broth), containing a Durham tube, is first used in the presumptive test to encourage the recovery and growth of potentially stressed coliforms in the sample. If harsher selective conditions are used, a deceptively low count may result. A tube containing both growth and gas is recorded as a positive result. It is possible for non-coliforms (Clostridium or Bacillus) to cause false positives in this medium and therefore all positive tubes are then inoculated into a more selective medium (Brilliant Green Lactose Broth or EC Broth) to begin the confirmed test.
The confirmed test medium effectively eliminates all organisms except true coliforms or fecal coliforms, depending upon the medium and incubation conditions. If a positive result is recorded in these tubes the completed test is begun by first streaking a loopful of the highest dilution tube which gave a positive result onto highly selective Eosin Methylene Blue (EMB) agar. After incubation, subsequent colonies are evaluated for typical coliform reactions.
The multiple tube fermentation test has the great disadvantage of taking 3-5 days to complete. If a municipality has a drinking water crisis, this is too long to wait. This has lead to development of faster, less complex tests. In the membrane filter technique 100 ml or greater of a test sample is passed through a filter with pores small enough to retain all bacteria in the sample. The filter is then placed on a selective medium that allows for the detection of coliforms. The advantages of this technique are the shorter time needed to complete the test (1 day vs. 3 to 4 days), its low cost, the higher accuracy in counting, since the colonies can be enumerated directly from the plate, and its simplicity. Disadvantages are that particulate samples (containing silt or other organic matter) quickly clog the filter, metals and phenols can stick to the filter inhibiting growth, and non-coliforms in the test sample may interfere with the formation of coliform colonies on the plate.
Recently, less complex tests for the detection of coliforms have become available. In the presence-absence test (P-A test), a large water sample (100 ml) is mixed with triple strength LLTB in a single culture bottle. Brom cresol purple is added as a pH indicator. If present, coliforms will ferment the lactose to acid and gas, turning the medium from purple to yellow. To detect coliforms and E. coli, the Colilert defined substrate test can be used. A 100 ml sample of water is mixed with a medium containing ortho-nitrophenyl-β,D-galactoside (ONPG) and 4-methyl umbelliferyl-β,D-glucoronide (MUG) as the only nutrients. If coliforms are present ONPG is metabolized, resulting in a yellow color. If E. coli is present, it will degrade MUG to a fluorescent product that can be detected by observation under long wave length UV-light. Both the P-A test and the Colilert defined substrate test are preliminary and any positive results will warrant further analysis of the offending sample.
The most probable number method of enumeration
In this experiment we will detect coliforms in a water sample using the multiple tube fermentation method. Enumeration of coliforms using this method involves inoculating multiple tubes with a 10-fold dilution series of the water sample and uses the most probable number (MPN) technique to estimate the population. To understand this technique, let us imagine preparing a 10-1 to 10-6 dilution series of a culture and inoculating 1 ml portions into tubes containing nutrient broth. After incubation, growth is observed in the tubes inoculated with the 10-1 to 10-5 dilutions, but not in the 10-6 tube. The number of organisms in the original sample is estimated to be less than 106, but greater than 105 bacteria per ml. By inoculating 3 or 5 tubes per dilution and using statistical analysis, a more accurate estimate of the bacterial concentration can be made. This is the basis of the MPN method. Realize that if a 10 ml solution contains 20 organisms, each 1 ml sample will probably not contain exactly 2 organisms. There will be some variation (0, 1, 2, 3 or 4 organisms/ml), but the total of ten 1 ml portions will add up to 20. This is why at lower dilutions, one tube inoculated with a certain dilution blank may show growth (receiving 1 or 2 organisms when inoculated) while other tubes inoculated with the same dilution do not (no organisms in the 1 ml). The number of organisms is assessed by counting the number of positives in the last three dilutions showing growth and then determining the MPN by following the directions on an appropriate table.
Period 1
Materials
Two samples for isolation of enterics. One can be any natural sample (for the general enteric isolation); the other should be a water sample from a lake or stream, especially a polluted area (for the water analysis).
1 flask of enteric enrichment broth (EE broth) 25 ml in 125 ml flask
15 tubes of Lactose Lauryl Tryptose Broth (LLTB)
2 99 ml dilution bottles (0.85% saline)
General Enteric Isolation
- If your sample is a solid, add 0.1-0.2 g to the flask of EE broth. If your sample is a liquid pipette 1 ml into the EE broth. Incubate the broth at 37°C on the shaker for 2-5 days.
- Water Analysis for Coliforms
- Label the two 99 ml dilution blanks 10-2 and 10-4. Shake the water sample vigorously and then dilute to 10-4 using the two 99 ml bottles.
- You will be testing a range of 10-0 to 10-4 for coliforms. Inoculate 1 ml of undiluted water sample into each of 3 tubes of LLTB. Label these 100. Inoculate 0.1 ml of the undiluted water sample into each of 3 tubes of LLTB, label these 10-1. Using the 10-2 dilution, inoculate 1 ml and 0.1 ml as above. Finally, take 1 ml of the 10-4 sample and inoculate it into the 3 remaining LLTB tubes.
- Incubate the tubes at 37°C for at least 48 hours.
Period 2
Materials
10-15 tubes of Brilliant Green Lactose Bile Broth (BGLB)
10-15 tubes of EC Broth
44.5°C water bath
1 plate of MacConkey's Agar Base + 0.2% glucose (MACG)
Figure 10.11. EE broth. Enteric enrichment broth after incubation. For your notebook describe the color of the broth and whether it is turbid.
Figure 10.12. LLTB. Lactose Lauryl Tryptose broth after incubation. Record the presence of growth and gas in the tubes. What is the physiological basis of the gas?
General Enteric Isolation
- Examine the EE broth. This medium is selective for enteric organisms due to the presence of bile salts and the dye, brilliant green. Prepare a Gram stain and verify the presence of Gram negative, non-spore forming, short rods.
- Streak the EE broth culture for isolated colonies onto MACG and incubate at 37°C for 1 day.
Water Analysis
- Examine the LLTB, looking for growth and gas in the tubes. Calculate the MPN from the pattern of positive (growth + gas) tubes observed. Use the MPN table Figure 10-X. This is the number of presumed coliforms.
- To continue the water analysis tests, inoculate a loopful of every positive tube of LLTB into a tube of EC Broth (to detect fecal coliforms) and into a tube of BGLB (to determine the confirmed number of total coliforms). Place the EC Broth tubes in a glass container (filled with 44.5°C water) and incubate in the 44.5°C water bath. Incubate the BGLB at 37°C. Continue incubation of both broths for at least 48 hours.
Figure 10.13. The MPN table. A table of statistical probability of cell number based upon growth in a three-tube dilution series. Use this table to estimate the number of microbes.
An MPN table for determining cell number from a three tube fermentation
| No of Tubes Positive In | No of Tubes Positive In | ||||||
|---|---|---|---|---|---|---|---|
| first | middle | last | MPN | first | middle | last | MPN¶ |
| 0 | 0 | 0 | <0.03 | 2 | 0 | 0 | 0.091 |
| 0 | 0 | 1 | 0.03 | 2 | 0 | 1 | 0.14 |
| 0 | 0 | 2 | 0.06 | 2 | 0 | 2 | 0.20 |
| 0 | 0 | 3 | 0.09 | 2 | 0 | 3 | 0.26 |
| 0 | 1 | 0 | 0.03 | 2 | 1 | 0 | 0.15 |
| 0 | 1 | 1 | 0.061 | 2 | 1 | 1 | 0.20 |
| 0 | 1 | 2 | 0.092 | 2 | 1 | 2 | 0.27 |
| 0 | 1 | 3 | 0.12 | 2 | 1 | 3 | 0.34 |
| 0 | 2 | 0 | 0.062 | 2 | 2 | 0 | 0.21 |
| 0 | 2 | 1 | 0.093 | 2 | 2 | 1 | 0.28 |
| 0 | 2 | 2 | 0.12 | 2 | 2 | 2 | 0.35 |
| 0 | 2 | 3 | 0.16 | 2 | 2 | 3 | 0.42 |
| 0 | 3 | 0 | 0.094 | 2 | 3 | 0 | 0.29 |
| 0 | 3 | 1 | 0.13 | 2 | 3 | 1 | 0.36 |
| 0 | 3 | 2 | 0.16 | 2 | 3 | 2 | 0.44 |
| 0 | 3 | 3 | 0.19 | 2 | 3 | 3 | 0.53 |
| 1 | 0 | 0 | 0.036 | 3 | 0 | 0 | 0.23 |
| 1 | 0 | 1 | 0.072 | 3 | 0 | 1 | 0.39 |
| 1 | 0 | 2 | 0.11 | 3 | 0 | 2 | 0.64 |
| 1 | 0 | 3 | 0.15 | 3 | 0 | 3 | 0.95 |
| 1 | 1 | 0 | 0.073 | 3 | 1 | 0 | 0.43 |
| 1 | 1 | 1 | 0.11 | 3 | 1 | 1 | 0.75 |
| 1 | 1 | 2 | 0.15 | 3 | 1 | 2 | 1.2 |
| 1 | 1 | 3 | 0.19 | 3 | 1 | 3 | 1.6 |
| 1 | 2 | 0 | 0.11 | 3 | 2 | 0 | 0.93 |
| 1 | 2 | 1 | 0.15 | 3 | 2 | 1 | 1.5 |
| 1 | 2 | 2 | 0.5 | 3 | 2 | 2 | 2.1 |
| 1 | 2 | 3 | 0.24 | 3 | 2 | 3 | 2.9 |
Figure 10.14. MACG plate after incubation. MacConkey's Agar plus glucose contains MacConkey agar base and glucose as the carbon source instead of lactose. All enterics will ferment the glucose to acid, and therefore form red colonies on the agar. Only select red colonies from this plate for futher study.
Figure 10.15. BGLB and EC tubes after incubation. Look for the presence of acid and gas in each tube. Note that tubes that were not inoculated last time would not have had growth, even if they were inoculated. Therefore, it is still possible to determine an MPN for BGLB and EC broth.
Period 3
Materials
2 slants of KIA
2 plates of EMB Agar
General Enteric Isolation
- Examine the MACG plate. Pick two potential enterics (How can this be determined? Think about the definition of an enteric and what is in MACG). Inoculate two different enteric isolates into KIA by taking a heavy loopful of culture, streaking the slant and then stabbing the butt of the agar. KIA is often used for preliminary identification - differentiation of potential enteric isolates and can be used as a screen to determine if the isolates are indeed Enterobacteriaceae.
- Incubate the KIA slants at 37°C for 2 days. (Place the KIA tubes in the rack up front; your instructor will incubate them.)
Water Analysis
- Examine the tubes of EC Broth and BGLB and calculate the MPN of confirmed fecal coliforms and coliforms in your sample. (You can perform this analysis because essentially you still have a 3 tube, 5 dilution, MPN series.) This constitutes the confirmed test for coliforms.
- Remove inoculum from the highest dilution of BGLB still showing growth + gas and streak for isolation onto a plate of EMB Agar. This constitutes the beginning of the completed test for total coliforms. Incubate at 37°C for 1 day.
- Repeat step 3 for fecal coliforms using the EC Broth and another plate of EMB agar. This is the beginning of completed test for fecal coliforms. Incubate at 37°C for 1 day. (Again, place the plates in the bin up front, your instructor will incubate them.)
Period 4
Materials
4 tubes of each of the following:
Lactose Fermentation Broth (with Durham tube)
Motility Indole Ornithine Medium (MIO)
Lysine Decarboxylase Broth (LDB)
Decarboxylase Control Broth (DCB)
Phenylalanine Agar Slants
Simmons Citrate Agar Slants
8 tubes of MR-VP broth
8 tubes of Sterile Mineral Oil
Figure 10.16. Typical reactions in KIA.
KIA is a multipurpose medium that tests four metabolic characteristics; Ability to ferment glucose, ability to ferment lactose, production of H2S, and production of gas. Using it, microbes can be classified into 5 groups.
- Non-enterics are unable to ferment glucose, and the KIA tube will remain orange (A).
- Those that have a yellow butt, but alkaline slant. (B) In this case, glucose is fermented, but lactose is not. The alkaline reaction observed in the slant is caused by the deamination of amino acids. H2S is not produced, there is no blackening of the medium from the formation of FeS.
- Those that form a yellow slant throughout the tube. (C) These organisms can ferment both lactose and glucose. The higher concentration of lactose in the medium causes a larger amount of acid production that overwhelms the alkaline reaction in the slant. H2S is not produced, there is no blackening of the medium from the formation of FeS.
- Those that have a yellow butt, but alkaline slant along with a blackening in the medium. (D) This is identical to #1 with the additional property of the production of 2S. This gas immediately reacts with iron in the medium forming FeS, which appears black.
- Those that form a yellow slant throughout the tube and have a blackening of the medium. (E)
General Enteric Isolation
- Check the KIA slants. Discard any isolate that is negative for glucose fermentation (e.g. red or orange butt) since it is not an enteric. Examine the slant and butt for net acid (yellow) or alkaline (red) reactions. The reddening of the slant is caused by aerobic deamination of amino acids, which results in a net alkaline reaction. A yellow butt is caused by the fermentation of sugars to acid. If a red slant is visible, the isolate is only able to ferment the relatively small amount of glucose in the medium. If the organism is able to ferment the greater concentration of lactose, enough acid will be produced to overwhelm the amino acid deamination in the slant causing a yellow color throughout the medium. Also, describe any blackening of the medium. This is caused by the production of H2S from thiosulfate by the test isolate. The H2S diffuses out of the cell and reacts with Fe in the medium to form FeS, a black insoluble compound. Enterics can be differentiated into 4 groups by KIA shown in Figure 10-15. Determine which group each isolate falls into.
- To help identify the genus of the bacteria isolated, use the verified enterics on KIA as a source of inoculum to perform the following tests.
- To confirm the fermentation of lactose, inoculate each isolate into Lactose Fermentation Broth. Incubate at 37°C for 2-5 days.
- To test each isolate for motility, indole production, and ornithine decarboxylation, inoculate a tube of MIO Medium by stabbing the center of the medium with the inoculating needle. Incubate for 1 day at 37°C.
- To test for phenylalanine deamination, inoculate the surface of a slant of Phenylalanine Agar. Incubate for 1 day at 37°C.
- To test for anaerobic lysine decarboxylation, inoculate each isolate into LDB and DCB. Overlay the tubes with sterile mineral oil. Incubate at 37°C for 2 days (or 30°C if incubation is to be for greater than 4 days)
- To test for utilization of citrate, inoculate the surface of a slant of Simmons Citrate Agar. Incubate at 37°C for 2-5 days.
- To investigate the type of end-products formed by fermentation, inoculate a tube of MR-VP Broth for the methyl red test. Inoculate another tube of MR-VP broth for the Voges-Proskauer test. Incubate at 37°C for at least 2 days.
Water Analysis
Figure 10.17. Growth on EMB. EMB is a highly selective medium for enteric bacteira. Eosin and methylene blue inhibibt the growth of many bacteria. Those that are capable of fermenting lactose will causes a drop in the pH of the medium. If lactose fermentation is rapid, colonies will have a green metalic sheen to them, forming coli-type colonies (A). If lactose fermentation is slower, colonies will be pink, with a dark center, forming fish-eye colonies (B). Non-lactose fermenting colonies will appear pink
- Examine the plates of EMB Agar and describe the colony morphology. Colonies of gram-negative lactose fermenting bacteria will show a relatively dark color. Coliform colonies can usually be separated into 2 types on EMB agar.
- coli-type - Colonies appear very dark, almost black when viewed directly and will have a green metallic sheen if observed by reflected light. The green sheen results from the rapid fermentation of lactose to acid, causing a low pH which precipitates methylene blue E. coli and those strains of Citrobacter which ferment lactose rapidly will form this type of colony.
- aerogenes-type - Colonies appear less dark. Normally a dark center is surrounded by a light-colored, mucoid rim. This characteristic formation is sometimes referred to as a fish-eye colony. Enterobacter and Klebsiella species often give rise to this type of colony.
- From each plate of EMB agar (coliforms and fecal coliforms) choose one coliform isolate for further analysis (including the isolate chosen in step 4). Inoculate the isolates into the medium used for identification of the general enteric isolates, as described in step 2.
Period 5
Materials
Dropper bottles of
Kovac's reagent
Methyl Red
FeCl3
α-naphthol (for Voges-Proskauer test)
40% KOH (for Voges-Proskauer test)
- Examine each reaction in the different media and perform each test as described below. It is most convenient to record results in tabular form, indicating a (+) or (-) for each test.
- Observe the Lactose Fermentation Broth and record whether the isolates are able to ferment lactose.
- Motility Indole Ornithine Medium. This medium is an excellent diagnostic tool since three tests can be performed by just one inoculation.
- Motility: The medium has a low concentration of agar (0.2%) which does not prevent motile organisms from migrating away from the stab line. Cloudiness throughout the medium is a positive result.
- Ornithine decarboxylation. Ornithine is an unusual amino acid (NH2CH2CH2CH(NH2)COOH) that can be decarboxylated by some enterics. MIO medium also tests for this. In a negative reaction, the glucose in the medium is fermented to cause a net acidic reaction and a yellow color. In a positive reaction the ornithine is decarboxylated anaerobically to produce a net alkaline reaction, which overcomes the acidic fermentation and causes a purple or bluish color in the medium.
- The indole reaction can also be tested in this medium. Tryptophan is provided in the medium and some enterics convert this to indole. The indole can be detected by addition of Kovac's reagent. The appearance of a red ring at the top of the medium signifies the production of indole. Add a dropperful of Kovac's reagent and then watch for a red ring.
- Lysine Decarboxylase (LDB) and Decarboxylase Control Broth (DCB). These two media are used to test for lysine decarboxylation.
- DCB (containing everything in LDB, except lysine) is necessary to verify that glucose in the medium is fermented to produce acid and for comparison to the LDB tube. Acid, from the fermentation of glucose, causes brom cresol purple in the medium to turn yellow. All enterics should show an acidic reaction in this medium. A negative reaction signifies that the isolate is a non-enteric.
- LDB. The presence of lysine decarboxylase is indicated if the tube has a color that is darker than the DCB. If observed, this is due to decarboxylation of lysine resulting in a net alkaline reaction. Record a positive test if the DCB tube is acidic (yellow) and the LDB tube is alkaline (purple or less yellow than the DCB tube)
- Phenylalanine Agar. One of the products from the deamination of phenylalanine is phenylpyruvic acid. This compound will react with Fe3+ to form a green complex. To test for phenylalanine deamination, add a few drops of FeCl3 and look for an immediate green color.
- Simmons Citrate Agar. If citrate is utilized as a carbon and energy source a net alkaline reaction (turning the medium blue) occurs due to the removal of citrate. Record this as a positive result.
- MR-VP Broth. Two general types of fermentation are used to differentiate enteric microorganisms, mixed acid fermentation and butanediol fermentation. The carbon source is initially converted to acid by both types of pathways. However, after 1-2 days the butanediol fermentation results in the production of alkaline products and a rise in pH. See Figure 9-2.
- To perform the methyl red test, gently layer 1/2 dropperful of methyl red onto the surface of a tube of MR-VP Broth. If a mixed acid fermentation has occurred, the medium will be acidic (pH < 4.4). and a red or pink color will be seen. If a butanediol fermentation has taken place, the medium will be more alkaline (pH > 6.2), producing a yellow color. Note that an equivocal reaction is also possible (6.6>pH>4.4) producing an orange color. Record a red or pink color (mixed acid fermentation) as a positive result
- The Voges-Proskauer (VP) test detects the production of acetoin. To perform the Voges-Proskauer test, add 12 drops of a-naphthol reagent and 4 drops of 40% KOH to a culture grown in MR-VP broth. Read results in about 10-30 minutes. If a red color is detected, this is a positive reaction and indicates the presence of the neutral end-product acetoin.
- Record all results in tabular form and compare them to the reaction of known organisms listed in Figure 10-1 below. Tentatively identify the genus of your isolates
Figure 10.18. Reactions in Fermentation Broths. A microbe capable of fermenting a sugar will lower pH because of the production of acidic end products. This figure depicts glucose fermentation broth. A negative reaction (A) shows turbidity (growth), but a purple color, since the pH has not changed. A positive reaction shows turbidity (growth) and a yellow color (B), due to the reaction of bromcresol purple with the acid produced during fermentation. A gas bubble in the Durham tube indicates the production of hydrogen gas. (C).
Figure 10.19. Reactions in MIO. MIO is a multipurpose medium that tests for motility, indole production and decarboxylation of ornithine. See the manual for details on these tests.
Figure 10.20. Reactions in LCB and DCB. Typical reactions observed for lysine decarboylation. Positive (A), negative (B). See text for details.
Figure 10.21. Reactions in Phenylalanine Agar. Some enterics are capable of deaminating phenylalanine forming phenylpyruvic acid. This will react with FeCl3 forming a green colored complex. Positive (A), Negative (B).
Figure 10.22. Simmon's citrate agar. Some enterics are capable of using citrate as a carbon source. In Simmon's citrate agar, this causes of the formationn of a blue metallic color. Positive (A), Negative (B)
Figure 10.23. The methyl red test. A major differentiating characteristic of enterics. Enterics with a mixed acid fermentation of sugars will drop the pH below 4.4, and appear red in this test when methyl red is added to the tube (A). Enterics with a butanediol fermentation of sugars will produce neutral end products later in growth, resulting in a increase in pH and will appear yellow in this test. (B). An equivocal reaction, between 4.4 and 6 is also possible and this has an orange color (C).
Figure 10.24. The Voges-Proskauer test. The Voges-Proskauer test detects the production of acetoin. α-naphthol reagent in an alkaline environment reacts with acetoin to form a red to pink color.
Differentiating tests for the Enterobacteriaceae
Figure 10.25. Reactions of various enterics in metabolic tests. A summary table listing the reactions of common enterics to all of the tests performed in this experiment. You may find a match here, but more likely you will need to use Bergey's Manual of Determinative Bacteriology to find your isolate.
| Primary Differentiating Tests | Secondary Differentiating Tests | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PA Deam | H2S Prod | Lac Ferm | Motility | Indole Prod | VP | Methyl Red | Sim Citrate | Lysine Decarb | Orn Decarb | |
| Citrobacter freundii | - | + | +/- | + | - | - | + | + | - | - |
| Edwardsiella tarda | - | + | - | + | + | - | + | - | + | + |
| Enterobacter aerogenes | - | - | +(w/gas) | + | - | + | - | + | + | + |
| Enterobacter cloacae | - | - | +(w/gas) | + | - | + | - | + | - | + |
| Escherichia coli | - | - | +(w/gas) | + | + | - | + | - | +/- | +/- |
| Hafnia alvei | - | - | - | [+] | - | [+]b | + | - | + | + |
| Klebsiella spp. | - | - | + (w/gas) | - | +/- | +/- | - | + | + | - |
| Kluyvera ascorbata | - | - | + | + | + | - | + | + | + | + |
| Morganella morganii | + | - | - | + | + | - | + | - | - | + |
| Proteus vulgaris | + | + | - | + | + | - | + | - | - | - |
| Proteus mirabilis | + | + | - | + | - | [+]b | + | +/- | - | + |
| Providencia spp. | + | - | - | + | + | +/- | + | + | - | - |
| Rahnella aquatilis | + | - | + | - | - | + | [+] | [+]b | - | - |
| Salmonella spp. | - | + | - | + | - | - | + | + | + | + |
| Serratia marcescens | - | - | - | + | - | + | - | + | + | + |
| Shigella spp. | - | - | - | - | +/- | + | + | - | - | +/- |